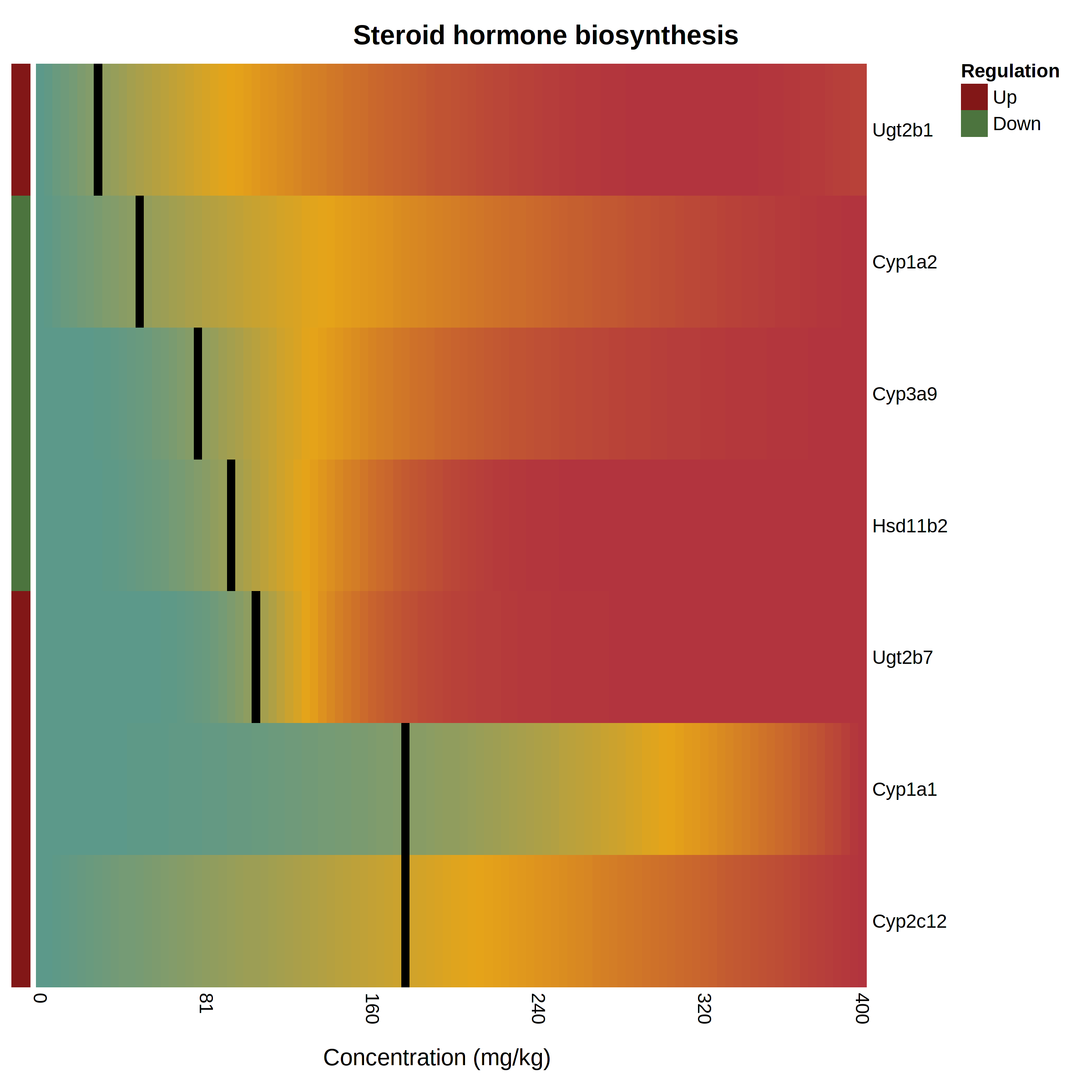
The pathway heatmap is an appealing visualization to clearly shows how the expression of each pathway gene compares to the others. It is generated when you click a pathway or gene set name in the “Gene Set Enrichment” panel at the result page. An example output is shown below The pathway heatmap values are calculated through a series of steps: The fitted model for each gene is evaluated across the range of doses in the uploaded data. The resulting modeled expression values are normalized
d2mvzyuse3lwjc.cloudfront.net/doc/en/Tutorial/imag
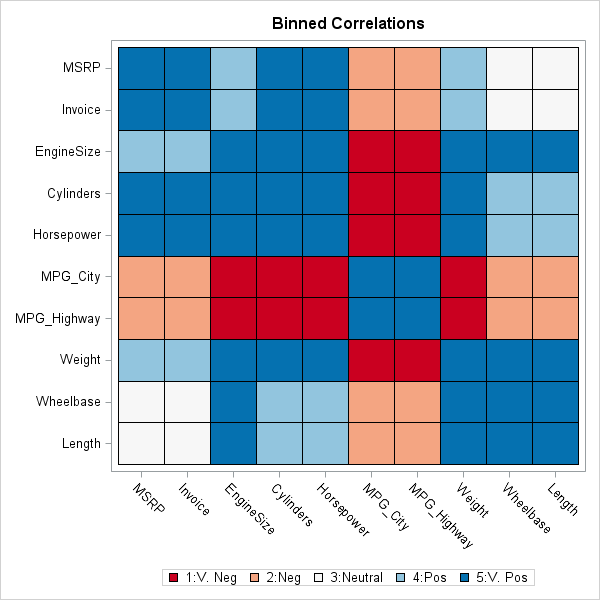
How to choose colors for maps and heat maps - The DO Loop
Support HCL colormaps in ComplexHeatmap - Zuguang Gu
How to choose colors for maps and heat maps - The DO Loop

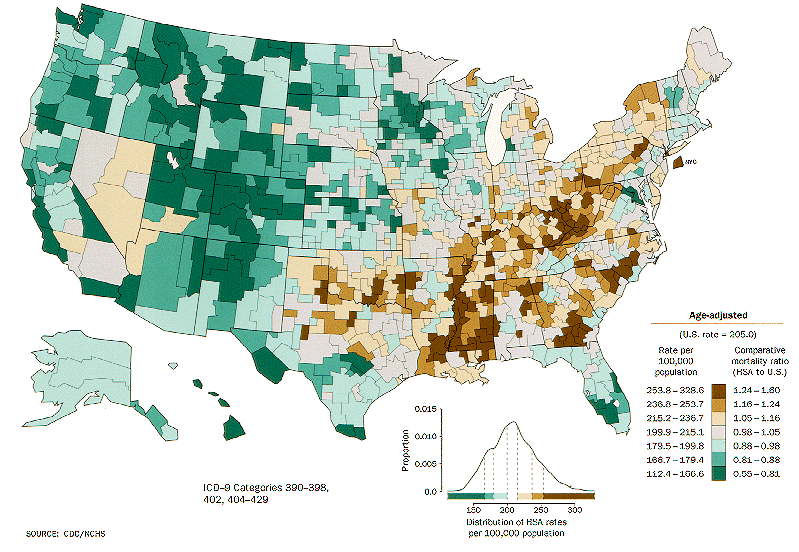
Scientific colour maps - s-Ink · accessible science graphics
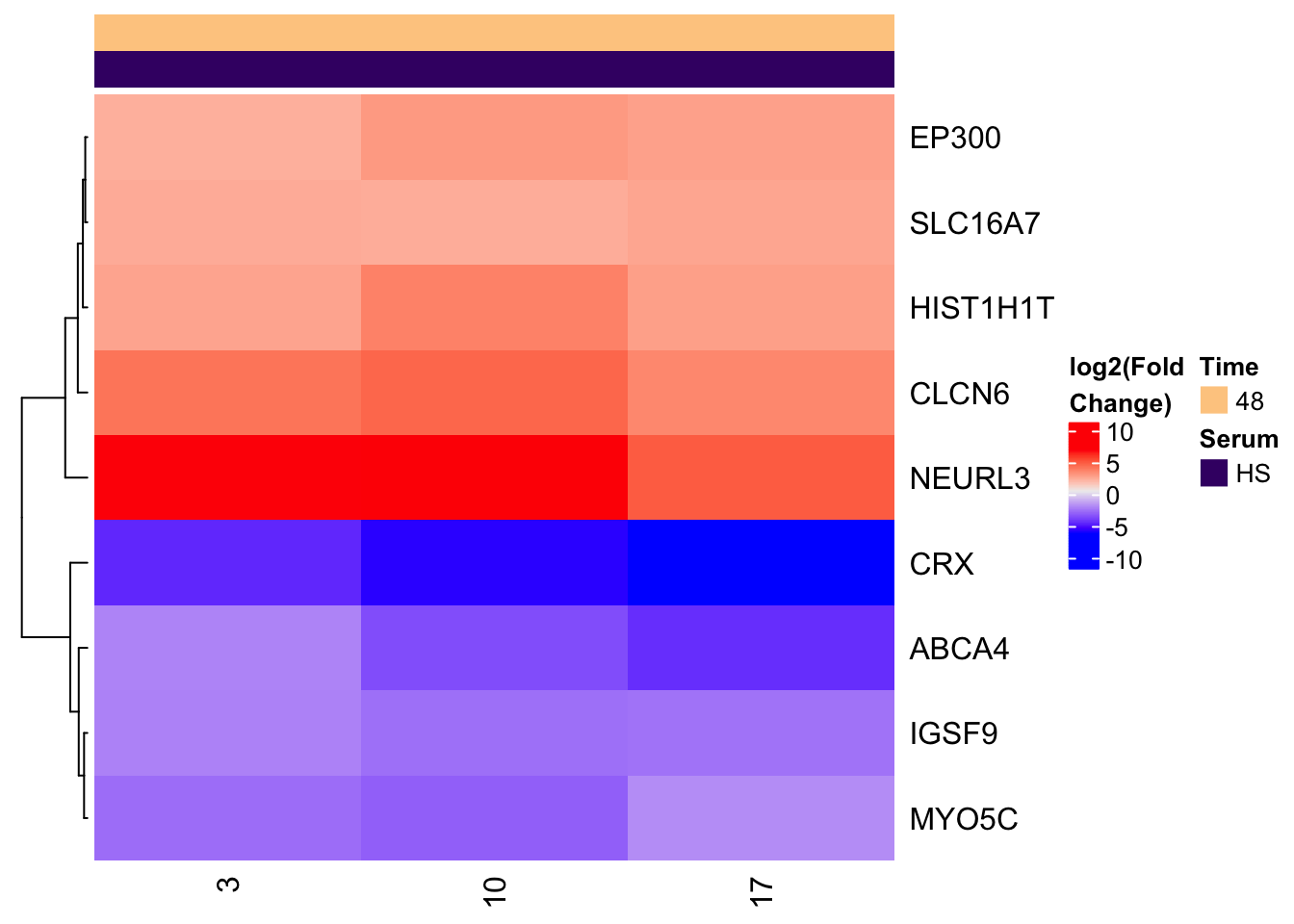
Let's Plot 6: Simple guide to heatmaps with ComplexHeatmaps
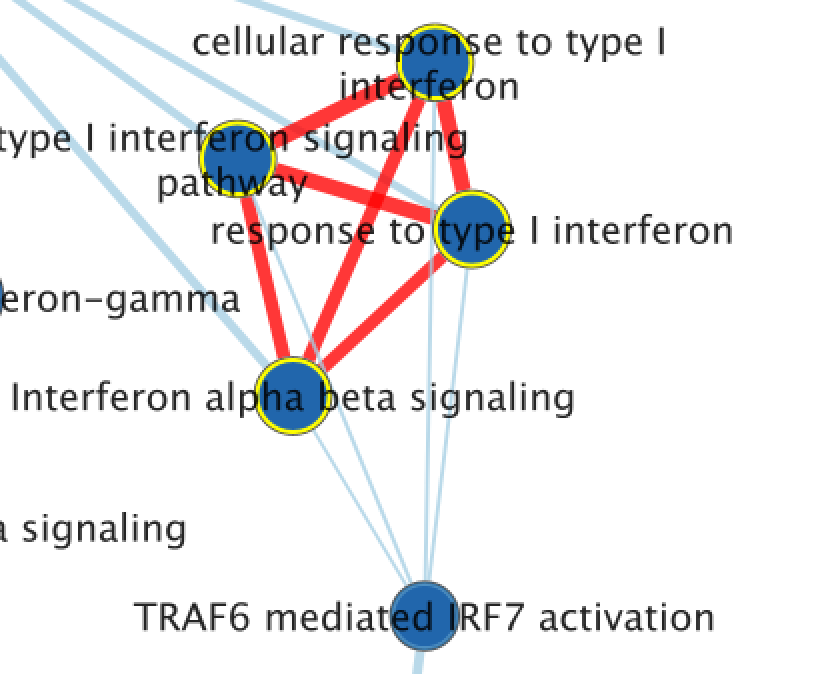
13A) Exploring the Table Panel heat map Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap

How to interpret heatmap ?

A Heatmap of-log10-transformed P-values for all 32 significant CP:KEGG
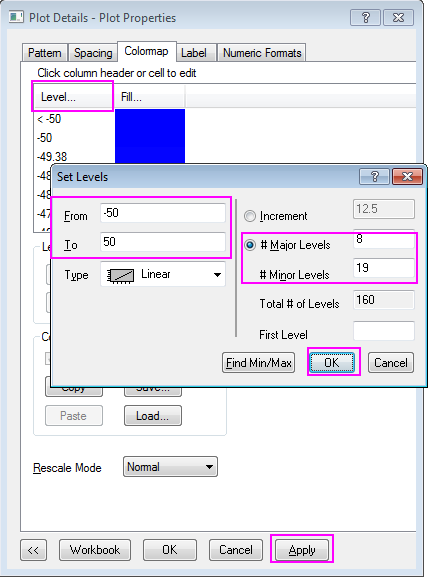
d2mvzyuse3lwjc.cloudfront.net/doc/en/Tutorial/imag
Heatmap - an overview – Flourish