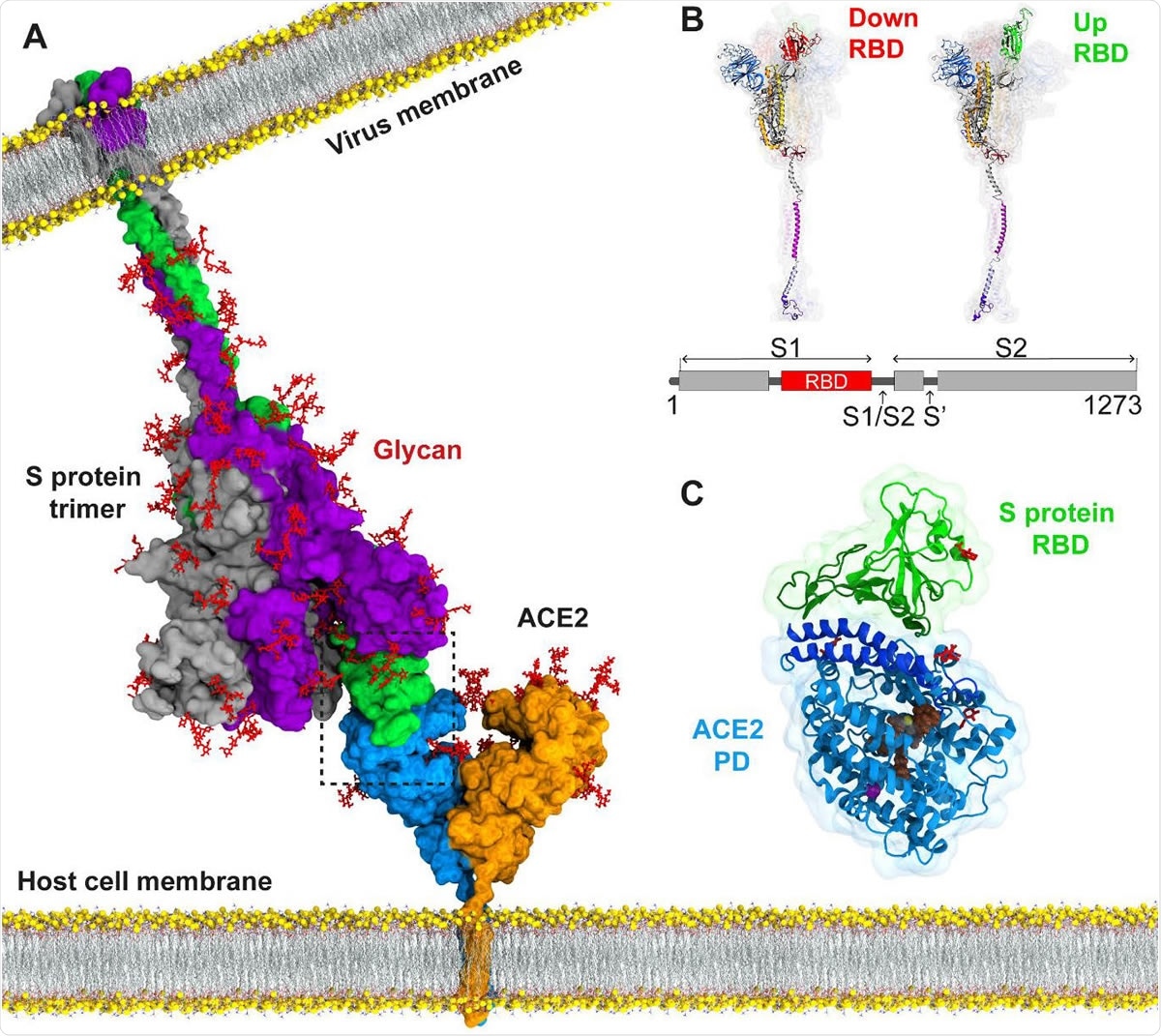
The current COVID-19 pandemic has spread throughout the world. Caused by a single-stranded RNA betacoronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is closely related to but much more infectious than the earlier highly pathogenic betacoronaviruses SARS and MERS-CoV, has impacted social, economic, and physical health to an unimaginable extent.
IJMS, Free Full-Text

Structural Modeling of the SARS-CoV-2 Spike/Human ACE2 Complex Interface can Identify High-Affinity Variants Associated with Increased Transmissibility - ScienceDirect

The effect of N-glycosylation of SARS-CoV-2 spike protein on the virus interaction with the host cell ACE2 receptor - ScienceDirect

Molecular dynamics simulation of S-nitrosylation of ACE2 a, Molecular
Molecules, -Text
The SARS-CoV-2 spike protein is vulnerable to moderate electric fields
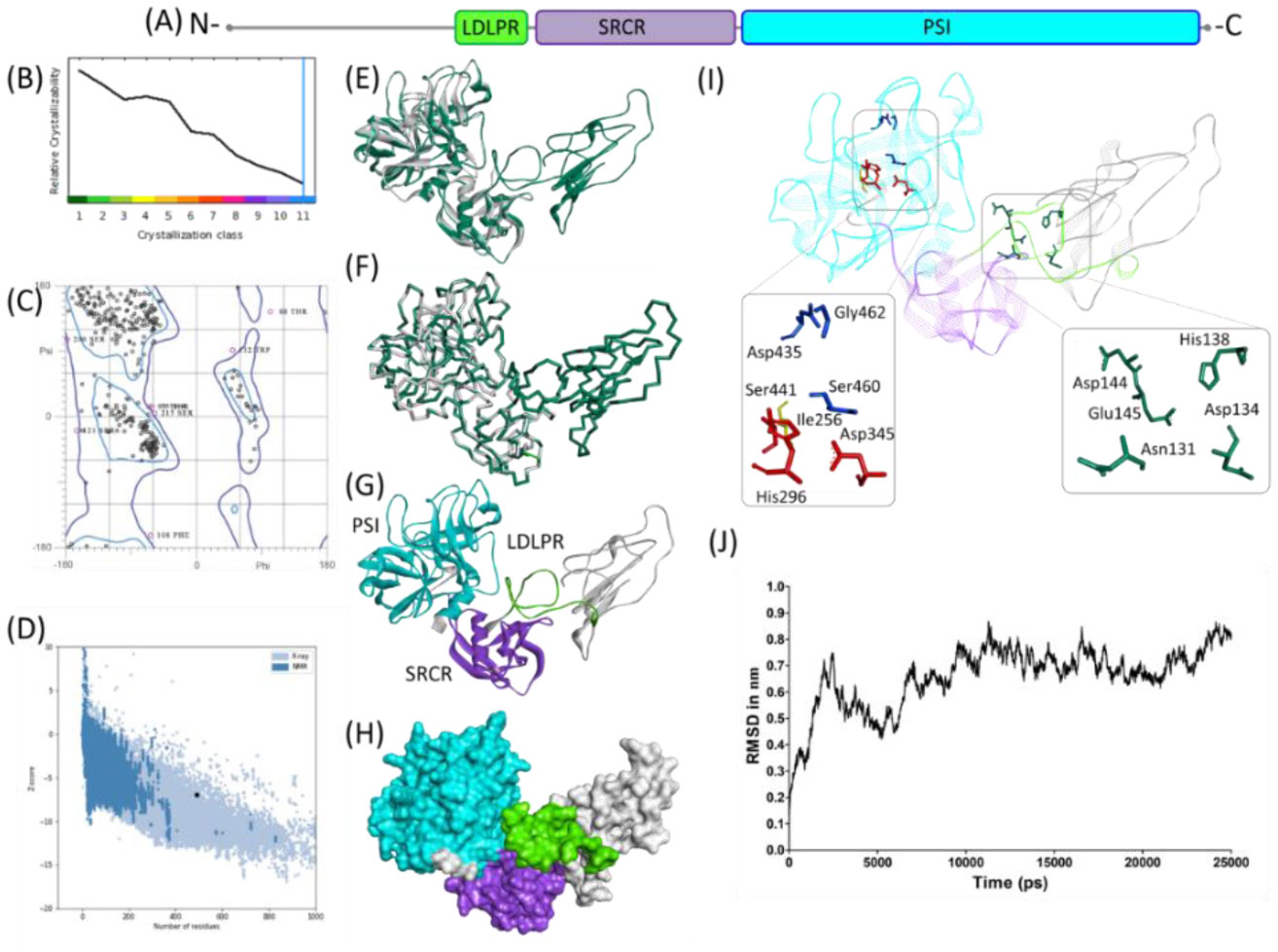
Molecular docking between human TMPRSS2 and SARS-CoV-2 spike protein: conformation and intermolecular interactions

In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin

Molecular dynamics simulations of RBD:ACE2 (as a reference) show

Cartoon depicting the interaction between the SARS-CoV-2 trimeric spike

Molecular dynamic simulations reveal detailed spike-ACE2 interactions
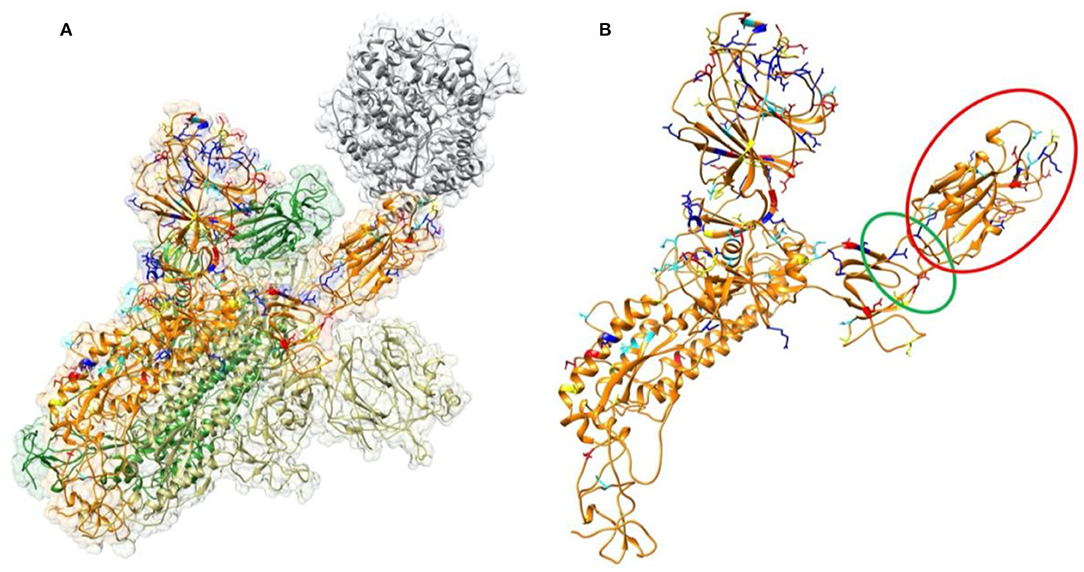
Frontiers Spike Proteins of SARS-CoV and SARS-CoV-2 Utilize Different Mechanisms to Bind With Human ACE2
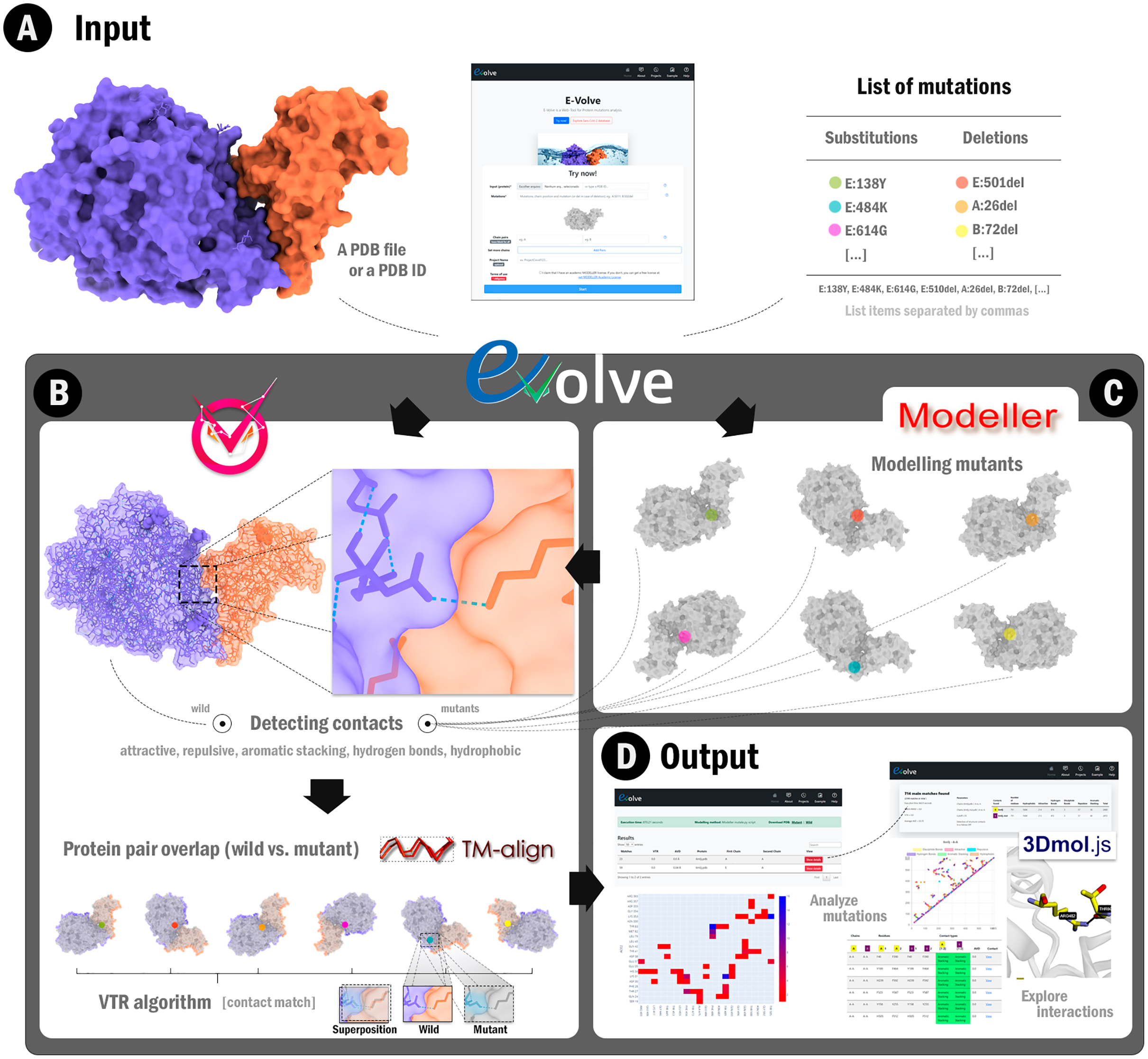
E-Volve: understanding the impact of mutations in SARS-CoV-2 variants spike protein on antibodies and ACE2 affinity through patterns of chemical interactions at protein interfaces [PeerJ]






